I’m currently working on a paper about how bacteria survive in extreme low temperature and salty environments. One thing that bacteria do when faced with these stresses is to produce protective chemicals called compatible solutes. Some microbes not only produce these chemicals but can also break them down for food and energy. Right now i’m interested in a polar microbe called Colwellia psychrerythraea 34H, which can break down these compatible solute compounds. Genes encoding the degradation of glycine betaine are found in this microbe, but not in any of its close relatives, suggesting that they were horizontally transferred into the genome at some point in the past.
The question I’m trying to answer now is: how far in the past? By looking at the ratios of the nucleotide bases guanine and cytosine (G+C) in the horizontally transferred DNA, and comparing them to those in the rest of the genome, we can start to answer this question.
The first thing I did was to look at the G+C content of each gene and compare it to that of the whole genome. In each case the gene is *much* more enriched in G+C compared to the rest of the genome (more than 2 standard deviations greater than the mean).
This provides more evidence that the genes were horizontally transferred into this bacterium. It also tells us that the donor bacterium had a G+C content greater than Colwellia 34H. What can that tell us? Well, DNA naturally accumulates mutations over time — this is an unavoidable fact. Because it takes three bases of DNA to code for one amino acid, there are redundancies in the Genetic Code, and so in many cases a change in the DNA sequence does not cause a change in the corresponding amino acid sequence. These changes are called ‘silent’ or ‘synonymous’ substitutions. As these changes accumulate over time in an organism’s genome, they trend towards an average value that is determined by the biochemical machinery of the cell, and the average value varies a lot among microbes (between about 20% and 80%). When a foreign gene is horizontally transferred into an organism that has a different G+C content, it will tend to ‘ameliorate’ over time. That is, the G+C content of the transferred gene will, over time, accumulate mutations that trend towards the G+C of the host organism.
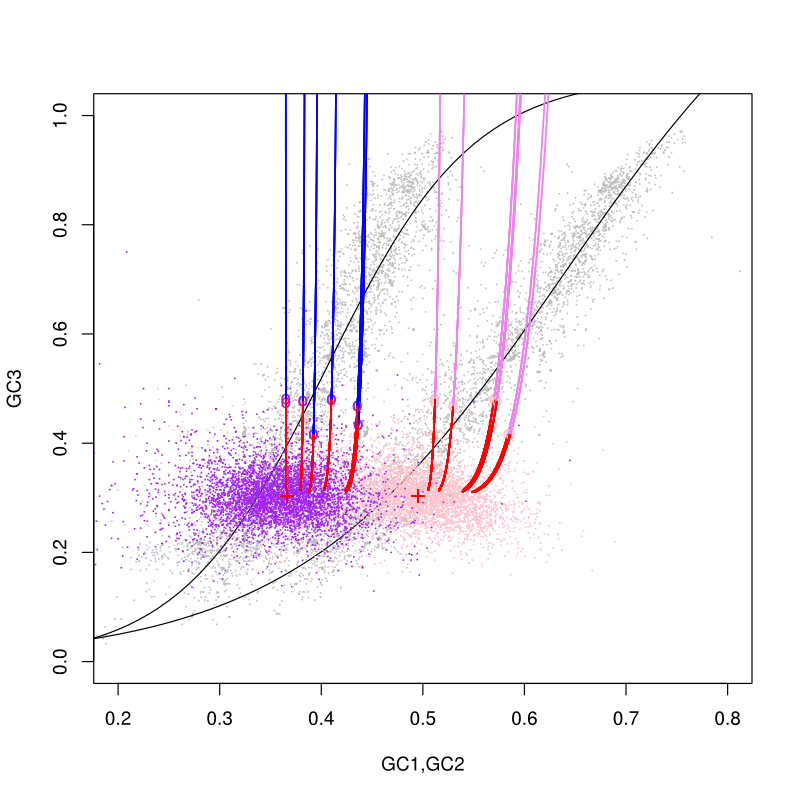
But how long does it take? It turns out that that depends on which codon position you look at, because some are more conserved in the genetic code than others. In general, the third codon position is more ‘degenerate’ than the first two and tends to accumulate synonymous mutations rapidly. The first position is the most conserved and tends to evolve the slowest. What this means is that we can look at the different rates of evolution at each position and make predictions about how the G+C content at that position will change over time — AND how it has already changed. A 1997 paper by Jeffrey Lawrence and Howard Ochman [1] laid out the theory behind this technique and wrote a program to calculate it. Unfortunately, the program only runs on ancient versions of windows (it was written for Windows 3.11!) — I tried running it in Wine on my Linux machine but it didn’t work properly. So I wrote some code [2] to do it in R instead. Here are the preliminary results, in the form of a pretty graph:
The curved lines are sigmoidal best-fit lines to the G+C contents of almost 4000 microbial genomes and plasmids (grey points), separate by codon position. The left curve is GC2 (evolves slowest), the right curve is GC1, and they are plotted against GC3 (evolves fastest) — rather than against whole genome G+C content, as in the original paper — to avoid biases associated with non-independence. The G+C content of each position in each gene in the host genome is also plotted as colored dots. The Lawrence and Ochman model is then used to iteratively ‘forward’ (red) and ‘reverse’ (blue or purple) ameliorate the G+C content of each horizontally transferred compatible solute degradation gene from it’s starting point (circles) to 1000 million years later (end point of each line), in time steps of 1 million years. As you can see, it takes a looong time to ameliorate these genes, but after a billion years we would expect these genes to have indistinguishable G+C content from their host, Colwellia 34H. Looking into the past, we pick the point at which they cross the best-fit curves as their ‘original’ G+C content and predict how many millions of years they have already been ameliorating in their host. That number provides the (approximate!) time since the horizontal gene transfer event occurred. In this case it appears that the genes were transferred between about 100 and 200 million years ago!
[2] GCamel software available at: http://github.com/rec3141/gcamel
Tags: ameliorate · bacteria · bioinformatics · evolution · gc content · github · horizontal gene transfer · science · software2 Comments




2 responses so far ↓
Did you publish this article in any journal? How can I cite your program?
Yes! This was published in Extremophiles https://link.springer.com/article/10.1007/s00792-013-0543-7